Statistical Inference of Transcription Cycle Parameters
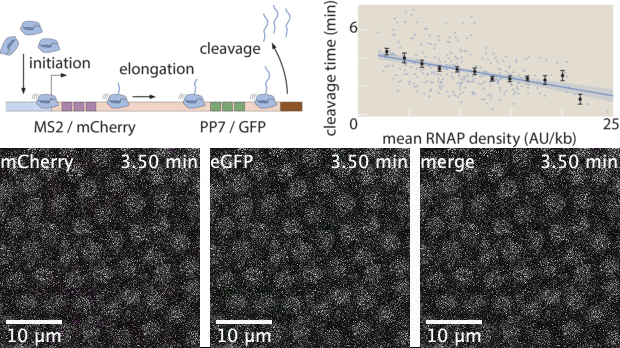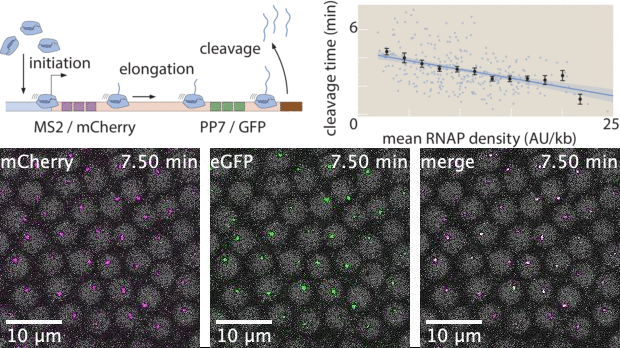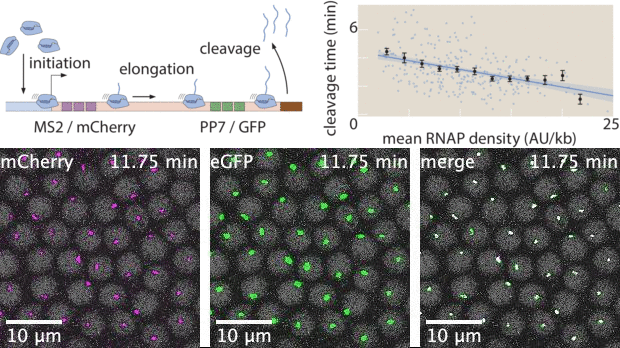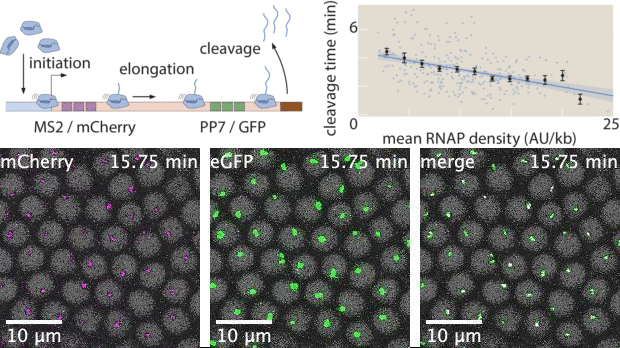
With live imaging tools such as MS2 or PP7, we can now image transcription at single-cell resolution in living organisms. However, these technologies yield large datasets that possess complex features both in space and in time. While heuristic analyses of live imaging measurements are common, the raw signals inside these datasets often possess much more information. Here, I used Markov Chain Monte Carlo (MCMC) to develop a statistical inference tool to extract effective transcription cycle parameters - mRNA initiation, elongation, and cleavage - from live imaging datasets. This method allows for quantitative estimation of kinetic processes involved in transcription at single-cell resolution, bridging the gap between raw experimental data and theoretical investigations that rely on statistics of biophysical parameters.